How to use PbsNRs
PbsNRs server contains three major functions: 1) Predict the activity of query compounds against different NR target; 2) Search the ligand files from built-in database; and 3) View the information of built-in database. Details description of each parts can be found as follows:
The accepted files of PbsNRs currently contains three types: 1) Smiles files (Paste Smiles to 1); 2) self defined molecular structures (Draw in 2); and 3) .Mol files (Paste Mol Block to 3), as shown in Figure 1.
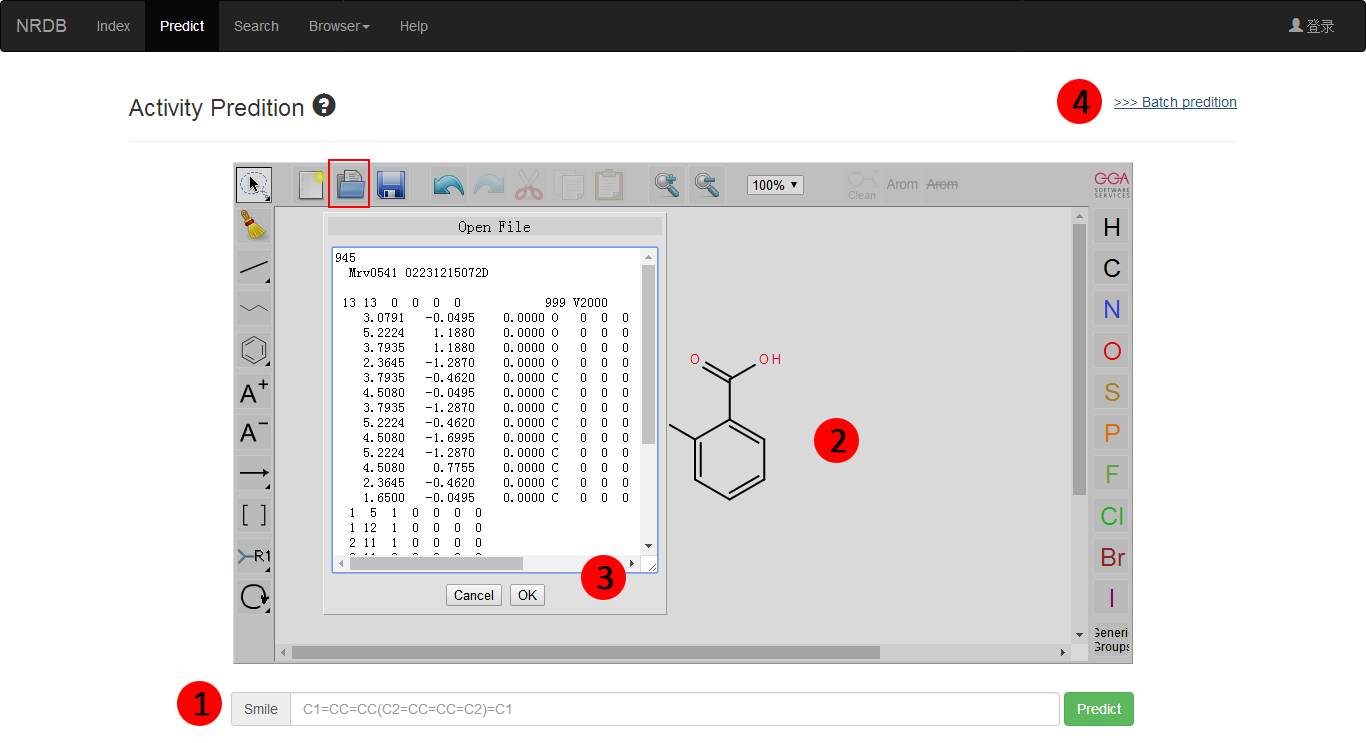
Figure 1. Input files for prediction.
The prediction results contains three parts:
1) Input Message, including the image and smile of the uploaded file;
2) The Murcko Scaffold of the inputted files;
3) Prediction Results, including the activity score for 30 NR target with target information. By click the NR ID, the 3-D structure of target protein can be viewed, also, the PDB id and Uniprot id were provided. For 11 NR targets within the experimental results, their protein-ligand activity information were also listed.

Batch upload is also encouraged and only smile files are accepted currently and the maximum upload files should be less then 100 (Figure 2). After upload the files, the server will automatically calculate the Activity score and derive molecular scaffold. The prediction results can be downloaded in .txt files.
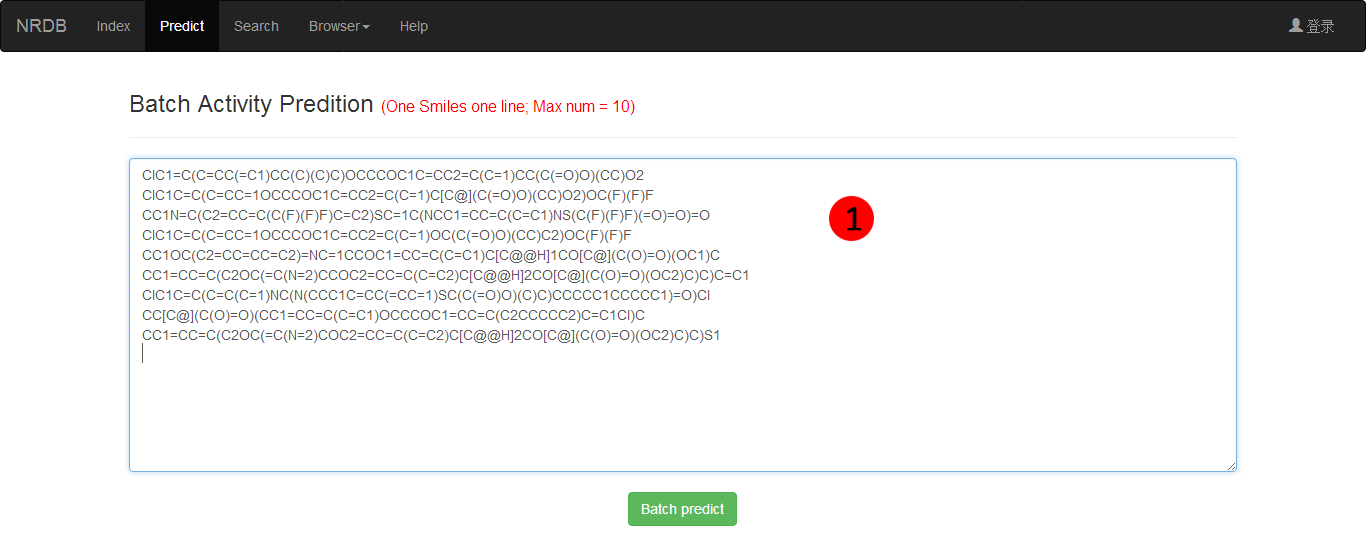
Figure 2. Batch queries for prediction.
Examples can be downloaded:
Smiles : 10 molecules.smiles
User can search the ligand files in our built-in database (Figure 3). Currently, PbsNRs provide two kind of search:
1) text search according to compound names;
2) similarity search according to structure similarities of molecules.
The results were listed in descending order based on similarity scores.
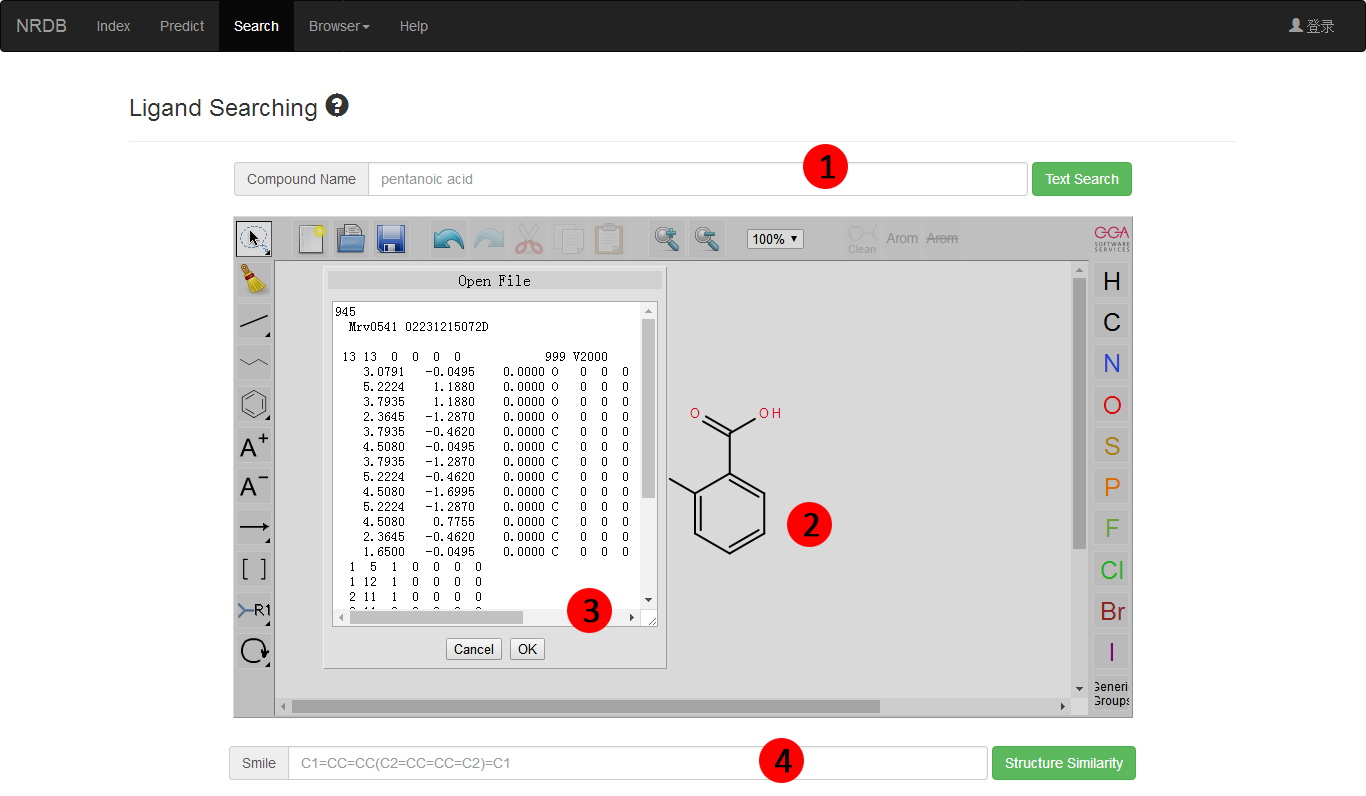
Figure 3. Search the ligand files in built-in database.
User can view the built-in database in four aspect: 1) information of available NR proteins; 2) information of NR ligands; 3) clustering of molecular scaffold; and 4) experimental activities in ascending orders.
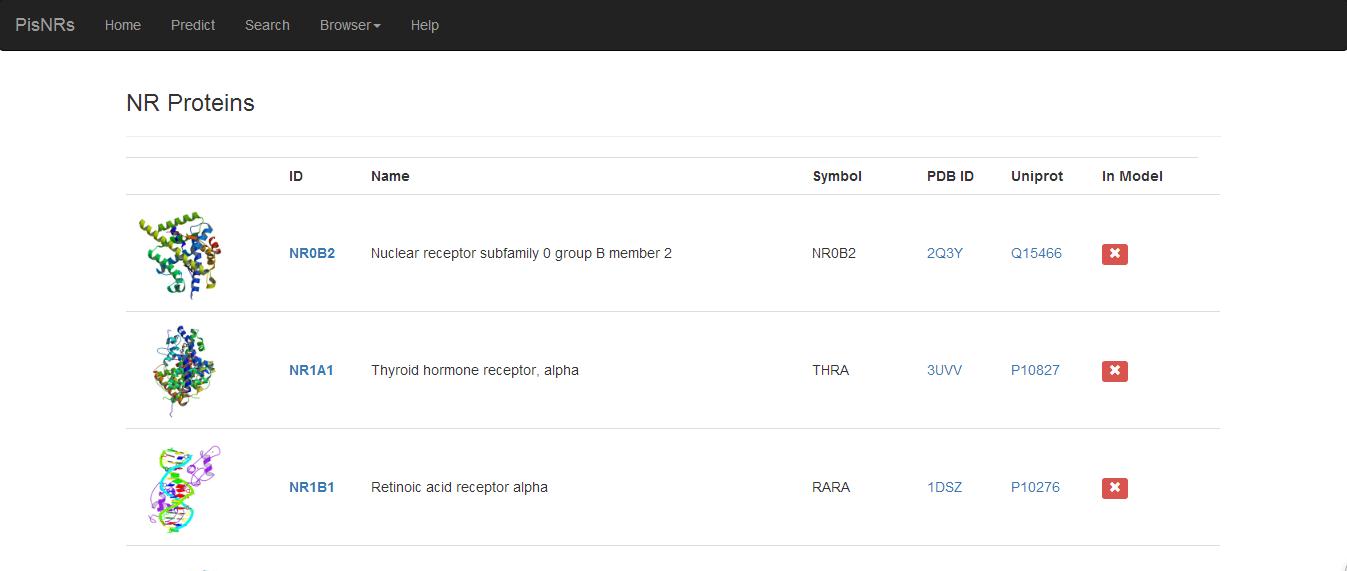
Figure 4. Browse NR proteins in built-in database.
Built-in database was collected from ONRLDB1.
Descriptors for proteins2,3 and ligands4
2. PDB Sequence Alignment Tools: Smith-Waterman
3. PDB Structure Alignment Tools: jFARCAT-flexible
Model created by Scikit-learn.5
5. Scikit-learn: Machine Learning in Python, Pedregosa et al., JMLR 12, pp. 2825-2830, 2011.
Web-based chemical structure editor : Ketcher6
1. One compound prediction : 1.130s
2. 10 compounds batch prediction : 7.233s
3. Text search : 0.440s
4. Strucutre search : 1.896s
You can predict activity of your compound by api : http://pisnrs.badd-cao.net/api/your smiles or http://www.badd-cao.net:1015/api/your smiles
It will return Murcko Scaffold and activity score of 30 NR proteins in JSON format.
Example : http://www.badd-cao.net:1015/api/CO[C@@H](CC1=CC=C(OCCC2=C(C)OC(=N2)C2=CC=CC=C2)C2=C1SC=C2)C(O)=O/
Python Package : https://pypi.org/project/pisnrs/
GitHub repositories : https://github.com/ddhmed/pisnrs
| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
|---|---|---|---|---|---|
| Linux | CentOS 7 | not tested | 60.2.2esr | n/a | n/a |
| MacOS | HighSierra | 73.0.3642.0 | 61.0.1 | n/a | 11.1(13605.1.33.1.2) |
| Windows | 10 | 71.0.3578.98 | 64 | 42.17134.1.0 | n/a |
pisnrs.badd-cao.net | 沪ICP备18014953号