1 Submission entrance
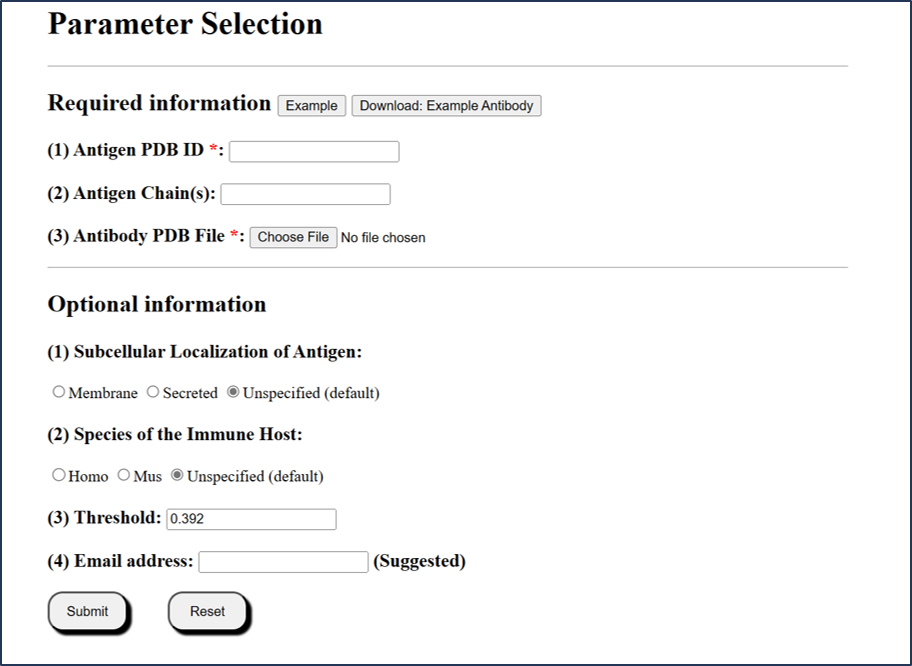
1.1 Submission mode 1 (via PDB ID)
Users can submit via an existing PDB ID (4 characters).
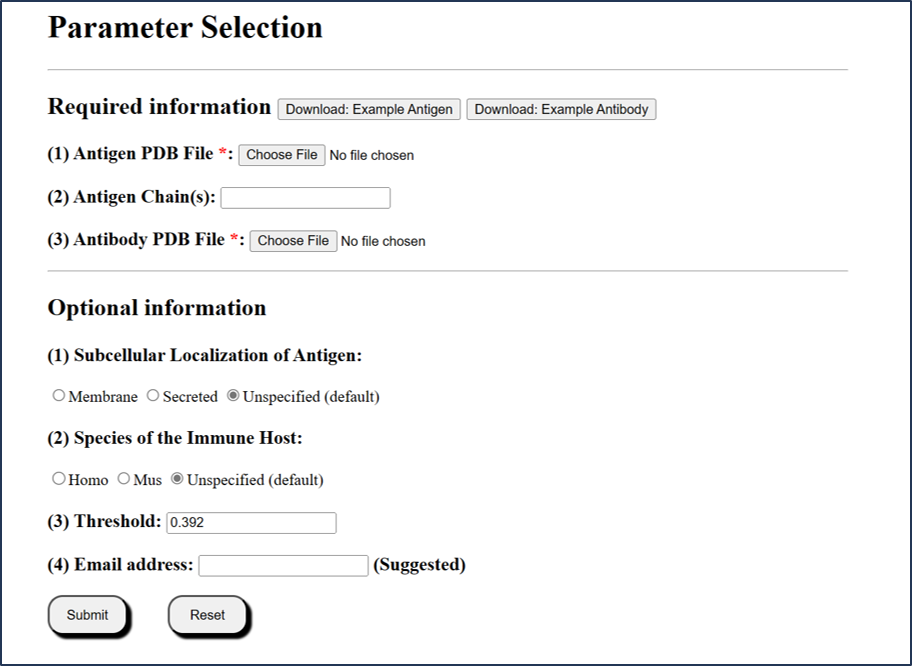
1.2 Submission mode 2 (via PDB file)
Users can submit local PDB files in PDB format.
For both above modes, please note that:
1) The chain(s) ID should be specified in each submission type.
2) If the chains are not specified, the whole structure will be calculated separately.
3) For local PDB file, be noted that chain column cannot be empty. Any chain ID with characters ("0-9" or "A-Z") is accepted.
4) For local PDB files, the filenames are only accepted with the following characters: "0-9", "A-Z" or "a-z".
5) Antibody structure is required in PDB format. If users only have sequence information of antibody, it is recommended to use SAbPred [1] and SWISS-Model [2] to build the structure file of the antibody.
[1] James Dunbar et al. Nucleic Acids Research, 2016, Vol. 44, W474-W478.
[2] Andrew Waterhouse, et al. Nucleic Acids Research, 2018, Vol.46, W296-W303.
1.3 Submission mode 3 (via Batch query)
Users can submit multiple PDB ID entries in batch query.
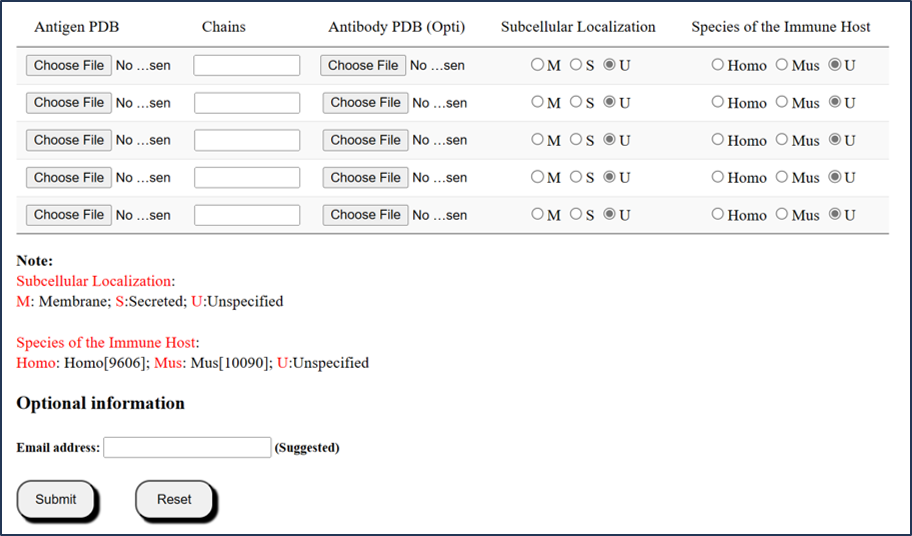
For batch query, SEPPA-mAb accepted no more than 5 files per batch.
1) User can upload less than 5 antigens in PDB format, with cognate antibody files in PDB format.
2) Note that, information of Subcellular Localization or Species of the Immune Host is optional based on the algorithm of SEPPA 3.0 (http://www.badd-cao.net/seppa3/).
3) Note that, epitopes prediction may take longer time than expected due to structure size and internet speed. Users are strongly encouraged to leave email address to receive results.
2 Example
Antigen File: 6CM3-E.pdb and Abntibody File: 6CM3-O-N.pdb
Example PDB ID: 6CM3 / Example file: 6CM3-E.pdb
Chain: E
Information selected:
1) Antibody info: 6CM3-O-N.pdb
2) Subcellular Localization of Antigen: Unspecified
3) Species of the Immune Host: Unspecified
4) Threshold: 0.392 (defaulted)
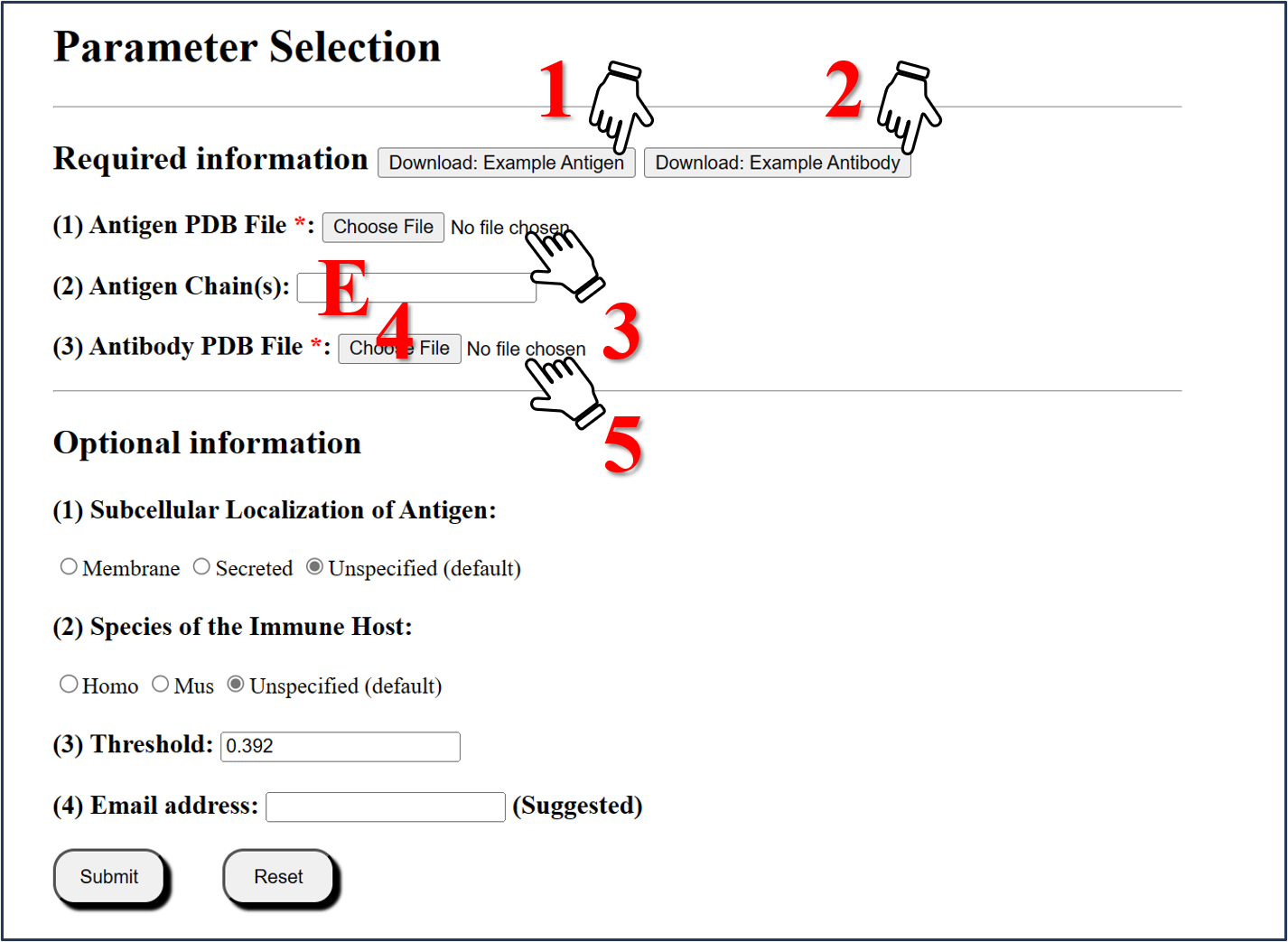
Step1: Download the example Antigen File
Click the Download: Example Antigen button to download "6CM3-E.pdb" file.
Step2: Download the example Antibody File
Click the Download: Example Antibody button to download "6CM3-O-N.pdb" file.
Step3: Upload Antigen structure
Select the "6CM3-E.pdb" to upload.
Step4: Selected Chain
Enter "E" for chain calculated.
Step5:Upload Antibody structure
Select the "6CM3-O-N.pdb" to upload.
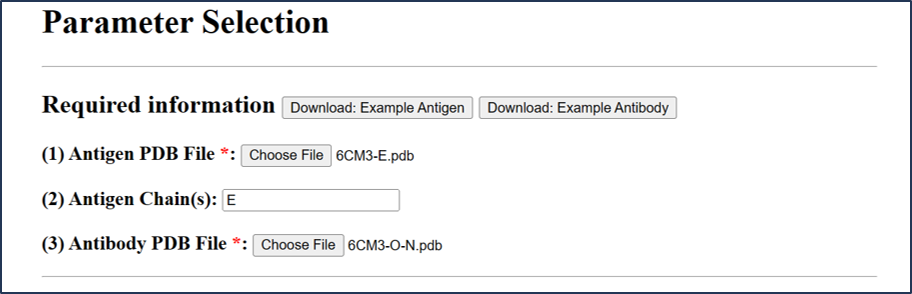
Submit information as above
2.1 Part I: Query information
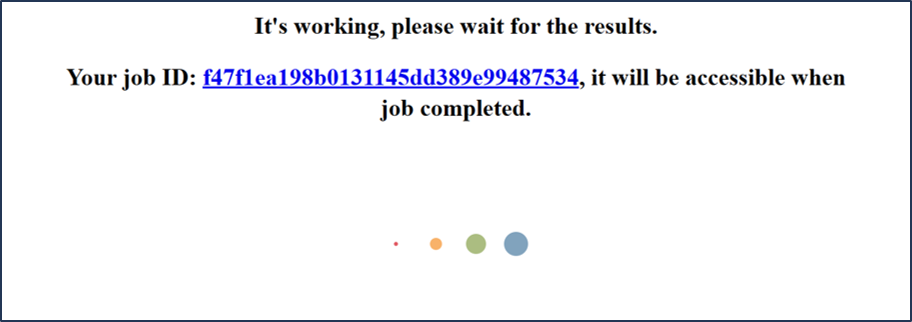
After clicking Submit button, users will enter a waiting page and a job-id is provided here.
Users can query historical Task via job-id from Query page.
2.2 Part II: summary of the prediction result
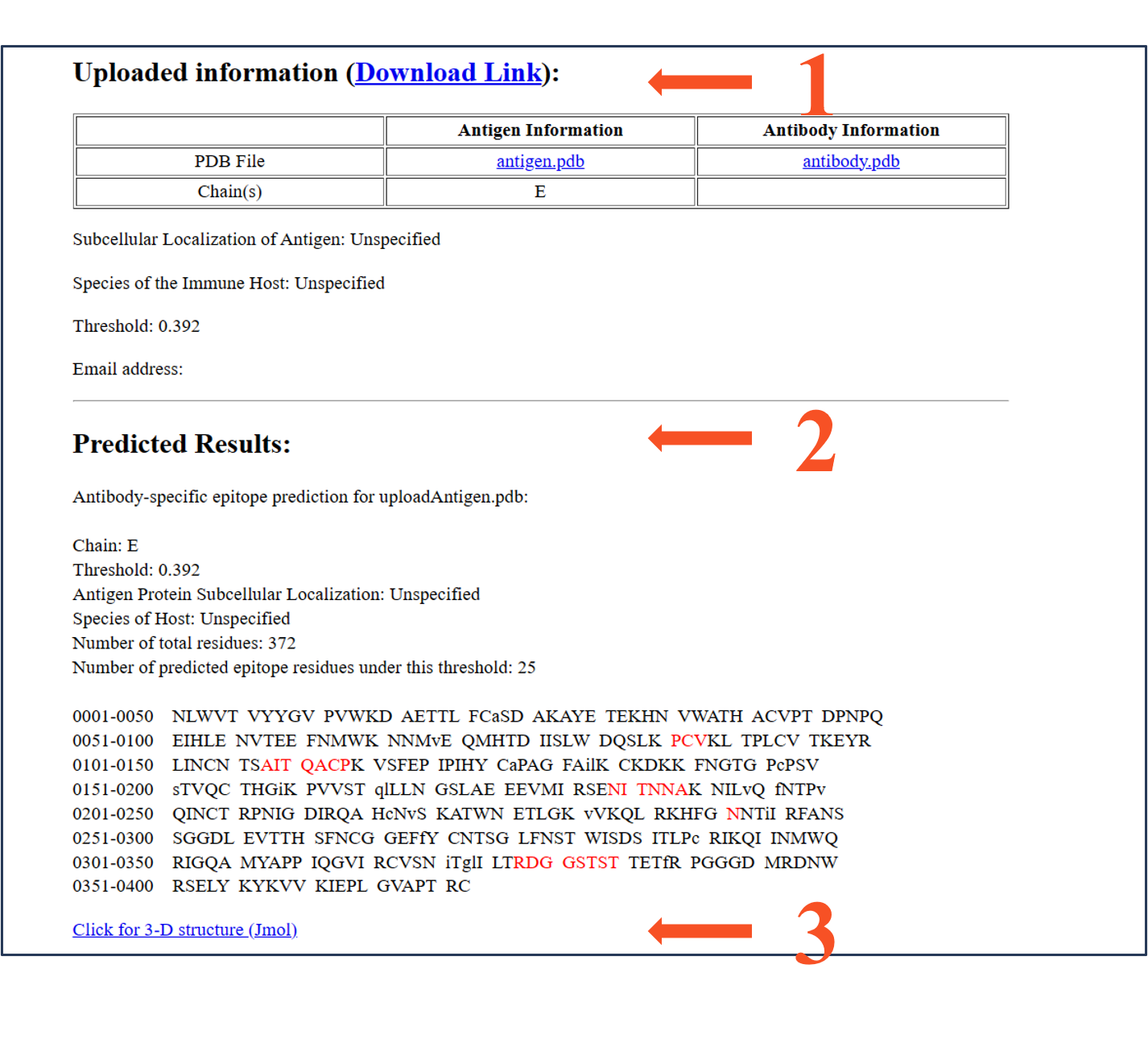
summary 1: Uploaded information
summary 2: Predicted Results
summary 3: visualization of the prediction result
summary 1: Uploaded information
Uploaded infomation: (Download Link):
Subcellular Localization of Antigen: Unspecified
Antigen Information
Antibody Information
PDB File
antigen.pdb
antibody.pdb
Chain(s)
E
Species of the Immune Host: Unspecified
Threshold: 0.392
Email address:
summary 2: Predicted Results
Antibody-specific epitope prediction for uploadAntigen.pdb:
Chain: E
Threshold: 0.392
Antigen Protein Subcellular Localization: Unspecified
Species of Host: Unspecified
Number of total residues: 372
Number of predicted epitope residues under this threshold: 25

The prediction result is displayed in a table as above.
Residues are listed sequentially.
The predicted epitope residues are highlighted in red.
The core residues are shown in lowercase, while the surface residues in uppercase.
summary 3: visualization of the prediction result
"Click for 3-D structure (Jmol)" link to a visualization page. See Part III for more detailed.
2.3 Part III: visualization of the prediction result
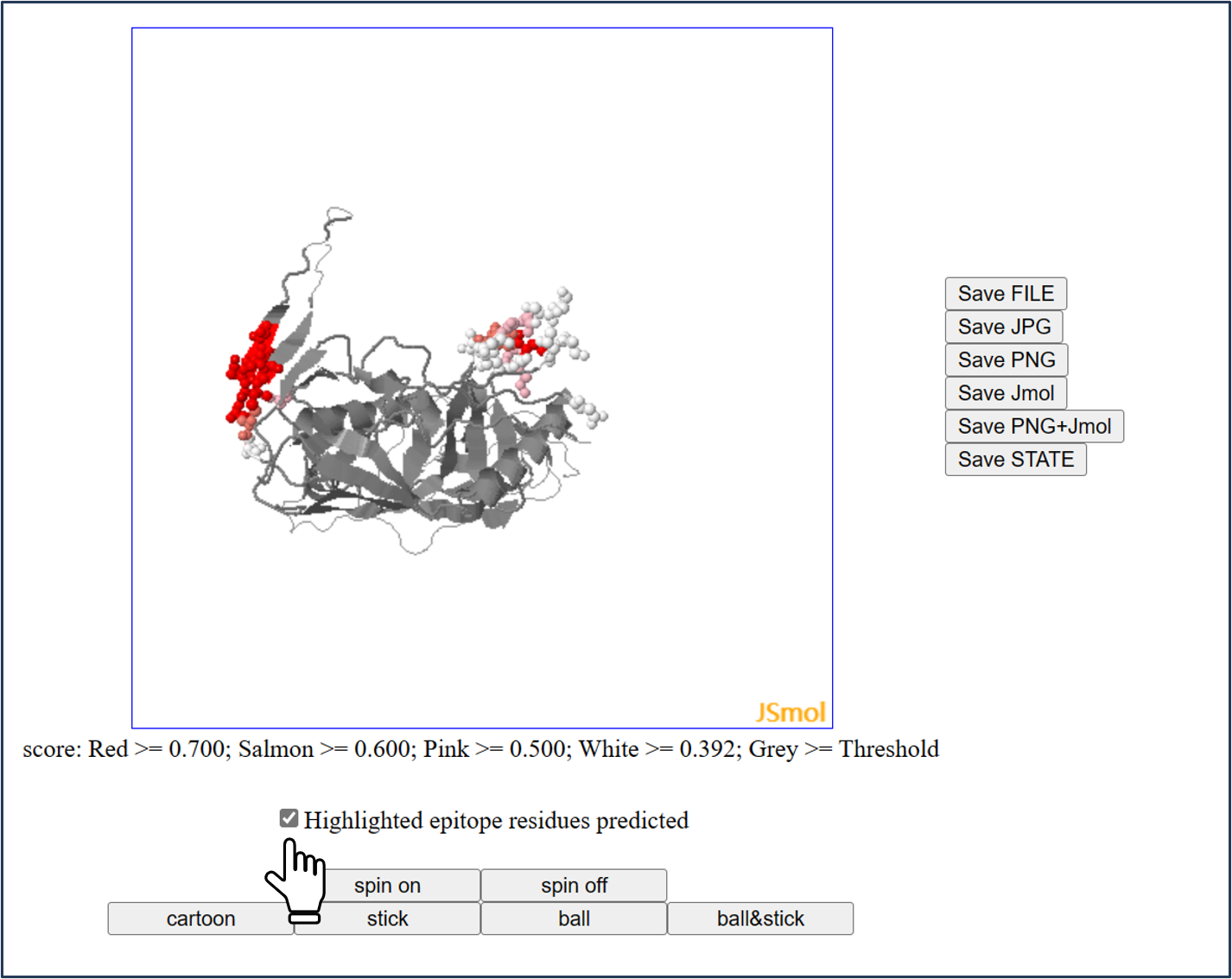
The 3D structure of antigen is showed by Jmol.
And the predicted epitopes will be labelled in color when users check the box.
All another operations are as same as Jmol.
* Different color shade indicates different epitope scores predicted:
-->Red: [0.7, 1.0]
-->Salmon: [0.6, 0.7)
-->Pink: [0.5, 0.6)
-->White: [0.392, 0.5)
-->Grey: [threshold, 0.392)
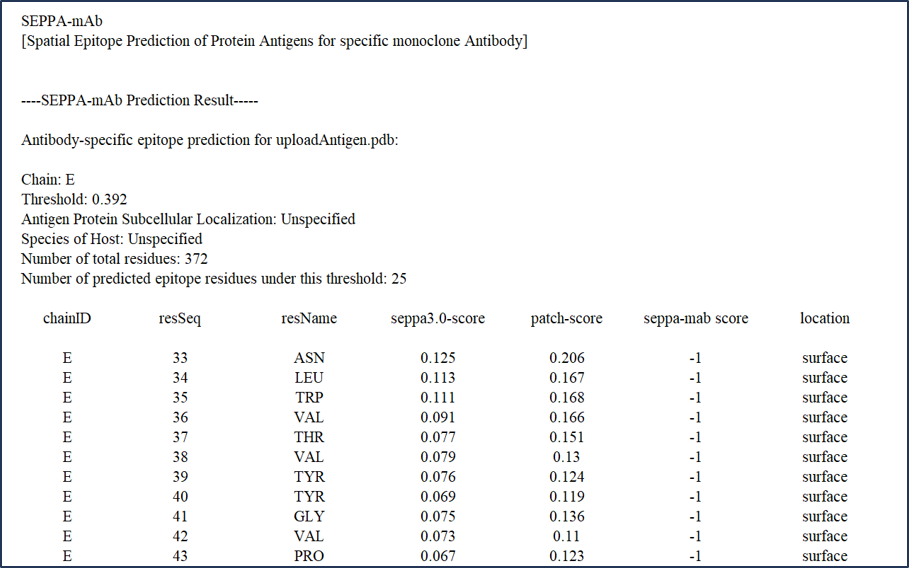
2.4 Part IV: score file of the prediction result (.txt) (via Download Link)
3 Browser compatibility
| System OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
| Linux | Ubuntu 17.10 | 71.0.3578.98 | 61.0.1 | n/a | n/a |
| MacOS | 10.13.6 | 107.0.5304.110 | 61.0.2 | n/a | 13.1.2 (13609.3.5.1.5) |
| Windows | 10.0.19044 | 108.0.5359.99 | 107.0.1 | 108.0.1462.42 | n/a |
ANNOUNCEMENT:
The residue solvent accessible areas are calculated with Naccess V2.1.1.
Hubbard SJ, Thornton JM. 'NACCESS', Computer Program.
Department of Biochemistry and Molecular Biology, University College London. 1993.
Jmol has been implemented into SEPPA-mab server.
Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/