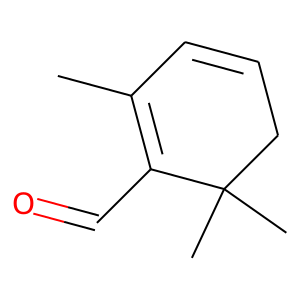
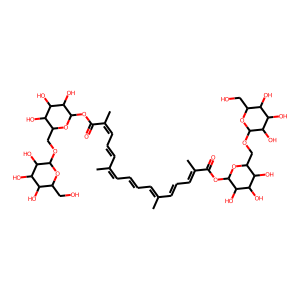
Target Description
Related Herbs (Total: 5)
| Herb id | Chinese Pin Yin | Chinese Character | English Name | Latin Name |
|---|---|---|---|---|
| H0182 | Luo Bu Ma Ye | 罗布麻叶 | NA | Apocyni veneti folium |
| H0095 | Gou Qi Zi | 枸杞子 | Chinese wolfberry fruit | Lycium chinense |
| H0818 | Zhi Zi | 栀子 | Cape jasmine fruit | Gardenia jasminoides |
| H0360 | Ren Zhong Bai | 人中白 | Human urine sediment | Homo sapiens |
| H0847 | Zi He Che | 紫河车 | Human placenta | Homo sapiens |